library(bbr)
library(here)
library(ggplot2)1 Introduction
For large or complex models, it is often useful to utilize parallel computing to make the models complete more quickly. This strategy effectively spreads the model execution over multiple CPU cores, so that independent parts of the fitting can be done simultaneously.
bbr makes it easy to parallelize your NONMEM models. This page demonstrates basic parallelization using bbr, as well as optimizing parallelization for a given model.
2 Tools used
2.1 MetrumRG packages
bbr Manage, track, and report modeling activities, through simple R objects, with a user-friendly interface between R and NONMEM®.
2.2 CRAN packages
ggplot2 A grammar of graphics.
3 Outline
When using bbr, you can easily parallelize your model via the threads value. This page shows how to parallelize a single model by using bbr::submit_model(..., .bbi_args = list(threads = <N>)). It also demonstrates using bbr::test_threads() to find the ideal value for threads for a particular model. As discussed below, the optimal number to pass to threads will vary significantly from model to model.
Note that this article only covers several basic use cases. For more detail on parallelizing with bbr, see Running NONMEM in Parallel: bbr Tips and Tricks. For an overview of using other bbr functionality for model development, see Model Management.
For a conceptual overview of parallelization, please see the Parallel Computing Intro and Grid Computing Intro articles in the Metworx Knowledge Base.
4 Set up
Load required packages and example model objects.
# you will need bbi >= 3.2.0 to use bbr::test_threads() as shown below
.v <- bbi_version()
if (.v == "" || package_version(.v) < 3.2) use_bbi() # this will install the latest versionModels 100 through 106 (shown in Model Management) are too simple to effectively run in parallel. In this example, we use model 200, which is intentionally more complex for the purpose of demonstrating parallel execution.
mod106 <- read_model(here("model", "pk", 106))
mod200 <- read_model(here("model", "pk-parallel", 200))
model_diff(mod106, mod200)@@ 1,3 @@@@ 1,3 @@<$PROBLEM From bbr: see 106.yaml for details>$PROBLEM From bbr: see 200.yaml for details$INPUT C NUM ID TIME SEQ CMT EVID AMT DV AGE WT HT EGFR ALB BMI SEX AAG$INPUT C NUM ID TIME SEQ CMT EVID AMT DV AGE WT HT EGFR ALB BMI SEX AAG@@ 6,5 @@@@ 6,8 @@$DATA ../../../data/derived/pk.csv IGNORE=(C='C', BLQ=1)$DATA ../../../data/derived/pk.csv IGNORE=(C='C', BLQ=1)<$SUBROUTINE ADVAN4 TRANS4>$SUBROUTINE ADVAN13 TOL=5~>$MODEL COMP(DEPOT)~>COMP(CENTRAL)~>COMP(PERPH)$PK$PK@@ 28,4 @@@@ 31,12 @@S2 = V2/1000 ; dose in mcg, conc in mcg/mLS2 = V2/1000 ; dose in mcg, conc in mcg/mL~>~>~>$DES~>~>DADT(1) = -KA*A(1)~>DADT(2) = KA*A(1) -(CL/V2)*A(2) -(Q/V2)*A(2) + (Q/V3)*A(3)~>DADT(3) = (Q/V2)*A(2) - (Q/V3)*A(3)~>$ERROR$ERROR@@ 51,5 @@@@ 62,6 @@0.05 ; 1 pro error0.05 ; 1 pro error<$EST MAXEVAL=9999 METHOD=1 INTER SIGL=6 NSIG=3 PRINT=1 RANMETHOD=P MSFO=./106.MSF>$EST MAXEVAL=9999 METHOD=1 INTER SIGL=6 NSIG=3 PRINT=1 RANMETHOD=P MSFO=./200.msf$COV PRINT=E RANMETHOD=P$COV PRINT=E RANMETHOD=P<$TABLE NUM IPRED NPDE CWRES CL V2 Q V3 KA ETAS(1:LAST) NOPRINT ONEHEADER RANMETHOD=P FILE=106.tab>$TABLE NUM CL V2 Q V3 KA ETAS(1:LAST) IPRED NPDE CWRES NOPRINT ONEHEADER FILE=200.tab~>$TABLE NUM CL V2 Q V3 KA ETAS(1:LAST) NOAPPEND NOPRINT ONEHEADER FILE=200par.tab
5 Submitting a model in parallel
There are several ways to control parallelization with bbr, but the simplest is to pass threads through .bbi_args, as shown below. The threads argument takes an integer, which tells it how many subprocesses to parallelize across. In practice, this will determine how many CPU cores it will use.
# execute a model, parallelized across 8 CPU cores
submit_model(mod200, .bbi_args = list(threads = 8))6 Optimizing the number of threads
Unfortunately, there is no easy way to know how many threads to parallelize a given NONMEM model across. You can expand the section below for more detail on that subject.
Additionally, it is important to understand that the benefits parallelization do not scale linearly. That is to say, doubling the number that you pass to threads will not make your model run twice as fast. As you scale up, you will begin to see diminishing returns and eventually you may even see it begin to take longer than it would have with fewer threads. (This is called “over-parallelizing” and is discussed further in Parallel Computing Intro.)
Given this nuance, you can use the bbr::test_threads() function to empirically estimate the ideal number of threads for a given model.
6.1 Testing number of threads empirically
The bbr::test_threads() function takes a model object, and a vector of integers passed to .threads, and does the following:
- Creates a copy of the input model for each value in
.threads - Optionally caps the number of iterations for the copied models
- Submits the copies to execute, each with one of the values passed to
.threads
test_mods <- test_threads(
mod200,
.threads = c(4, 8, 16, 32, 64, 96),
.cap_iterations = 100
)Once you have submitted your test models, you can pass the returned list of model objects to bbr::check_run_times(). This will hold your console until all the test models have finished running, and then return a simple tibble with the results from the test runs.
res <- check_run_times(test_mods) # holds console until all test models finish
res| run | threads | estimation_time |
|---|---|---|
| 200_4_threads | 4 | 66.97 |
| 200_8_threads | 8 | 36.18 |
| 200_16_threads | 16 | 24.22 |
| 200_32_threads | 32 | 14.66 |
| 200_64_threads | 64 | 13.74 |
| 200_96_threads | 96 | 15.29 |
6.2 Plot to visualize run times
Often, the table above is simple enough to give you an answer, but it is also easy to visualize these results.
ggplot(res, aes(x = threads, y = estimation_time)) +
geom_point() +
geom_line() +
scale_x_continuous(breaks = res$threads, minor_breaks = NULL) +
xlab("Number of Threads") +
ylab("Estimation Time (in seconds)")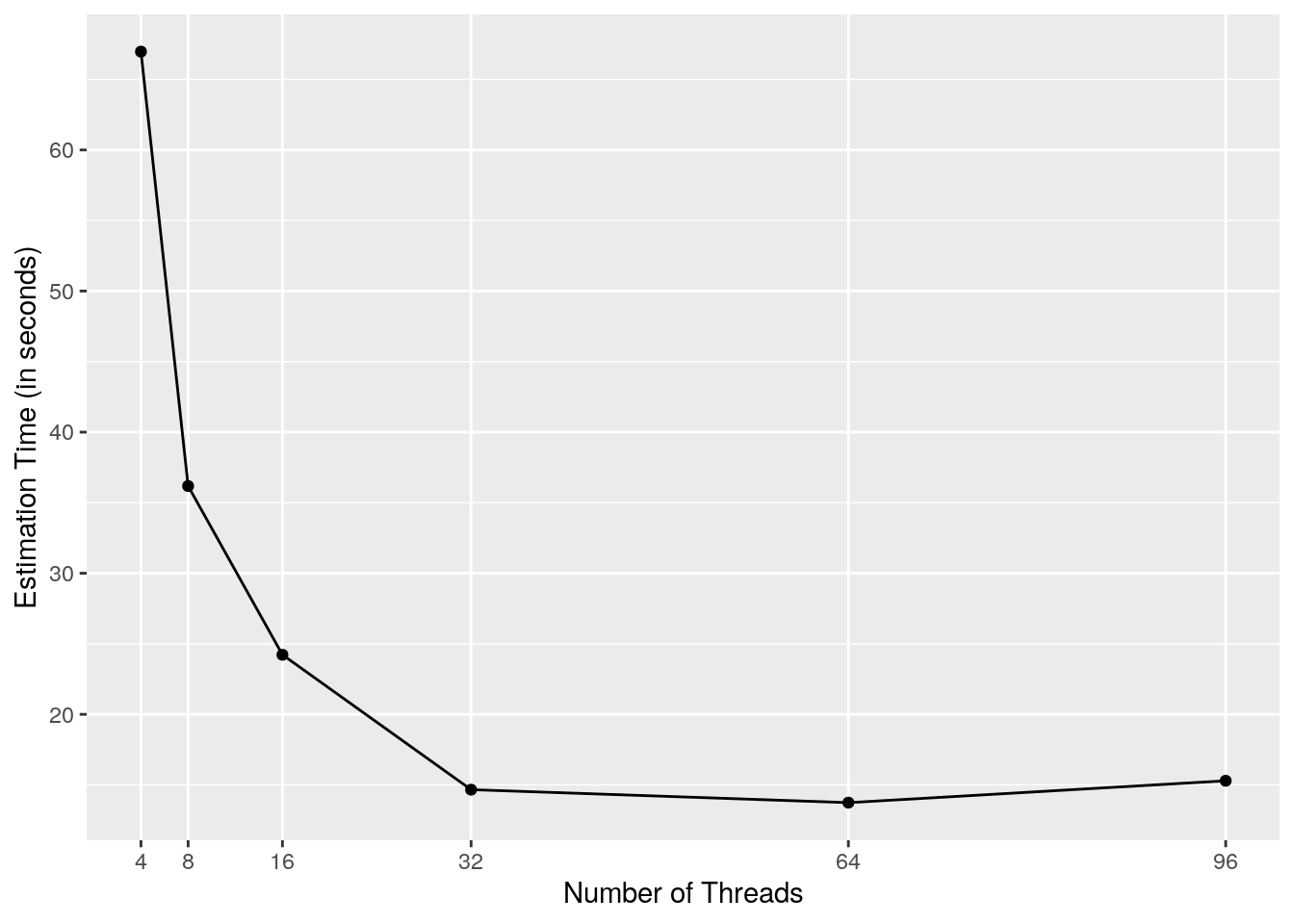
The plot above highlights an important point that we touched on above: the benefits parallelization do not scale linearly. Notice how we see significant (though not linear) speed gains when increasing from 8 to 16, or 16 to 32, threads. However, increasing to 64 threads provides very limited improvement, and increasing to 96 threads actually makes the model take more time.
Consider that, if you are using some kind of auto-scaling infrastructure (discussed in the next section), you are likely paying money for each CPU core that you request. In this light, something like 32 threads looks ideal for this model.
6.3 Auto-scaling in Metworx
By default, bbr will submit your job to run on an SGE grid. This is ideal for systems like Metworx, which will auto-scale compute resources based on the amount of work that has been submitted. If you are using Metworx (or another system using SGE) you can view your submitted jobs with the qstat utility.
system("qstat -f")############################################################################
- PENDING JOBS - PENDING JOBS - PENDING JOBS - PENDING JOBS - PENDING JOBS
############################################################################
7 0.60500 Run_200_96 <user> qw 01/26/2023 13:11:11 96
6 0.57022 Run_200_64 <user> qw 01/26/2023 13:11:11 64
5 0.53543 Run_200_32 <user> qw 01/26/2023 13:11:11 32
4 0.51804 Run_200_16 <user> qw 01/26/2023 13:11:11 16
3 0.50935 Run_200_8_ <user> qw 01/26/2023 13:11:11 8
2 0.50500 Run_200_4_ <user> qw 01/26/2023 13:11:11 4 When you first call test_threads(), you will likely see something like this, indicating that your jobs have been submitted to the queue and are waiting for the appropriate resources to be launched by the auto-scaling grid. The right-most column indicates the number of CPU cores that a given job is requesting.
If you run qstat -f again, several minutes later, you will see compute nodes begin to come online and start executing these jobs. Once all of the jobs have finished, the compute nodes will watch the queue for 15 minutes before scaling back down (i.e. turning themselves off) if they have no additional work.
6.4 Clean up test models
Once you have a settled on an appropriate number of threads to use, you will likely want to clean up the test models that test_threads() created. Pass the list of models (that was returned from test_threads()) to bbr::delete_models(). This will clean up all on-disk files from these models.
delete_models(test_mods)7 Other resources
The following script from the Github repository is discussed on this page. If you’re interested running this code, visit the About the Github Repo page first.
- Runnable version of the examples on this page:
nonmem-parallel-demo.R