head(data)1 Introduction
The pmforest package can be used to visualize the covariates of interest in your analysis by creating grouped displays of point estimates and variability ranges for any kind of continuous data.
2 Example data
To illustrate this package, we created some example data looking at dose adjustments for renal impairment. Here we’ve calculated AUCss at different doses for subjects with varying levels of renal function.
ID RFf dosef type CL AUC
<int> <fctr> <fctr> <fctr> <num> <num>
1: 1 Normal function 10 mg original 2.536364 3.942651
2: 2 Normal function 10 mg original 1.555193 6.430070
3: 3 Normal function 10 mg original 2.579118 3.877295In one set of simulations, a standard set of doses were evaluated regardless of renal function (Original) while another simulation set implemented a simple dose adjustment scheme for subjects with moderate or severe renal impairment (Adjusted).
3 Forest plots
To display the central tendency and variability in these data graphically using the pmforest package we follow a two-step procedure.
3.1 Step 1: summarize the data
We call summarize_data() to create data summaries that will be plotted.
adjusted <- filter(data, type=="adjusted")
summ1 <- summarize_data(
data = adjusted,
value = "AUC",
group = "RFf",
group_level = "dosef"
)This function provided by pmforest creates a summary data frame that is in the correct format for processing by plot_forest() in the next step.
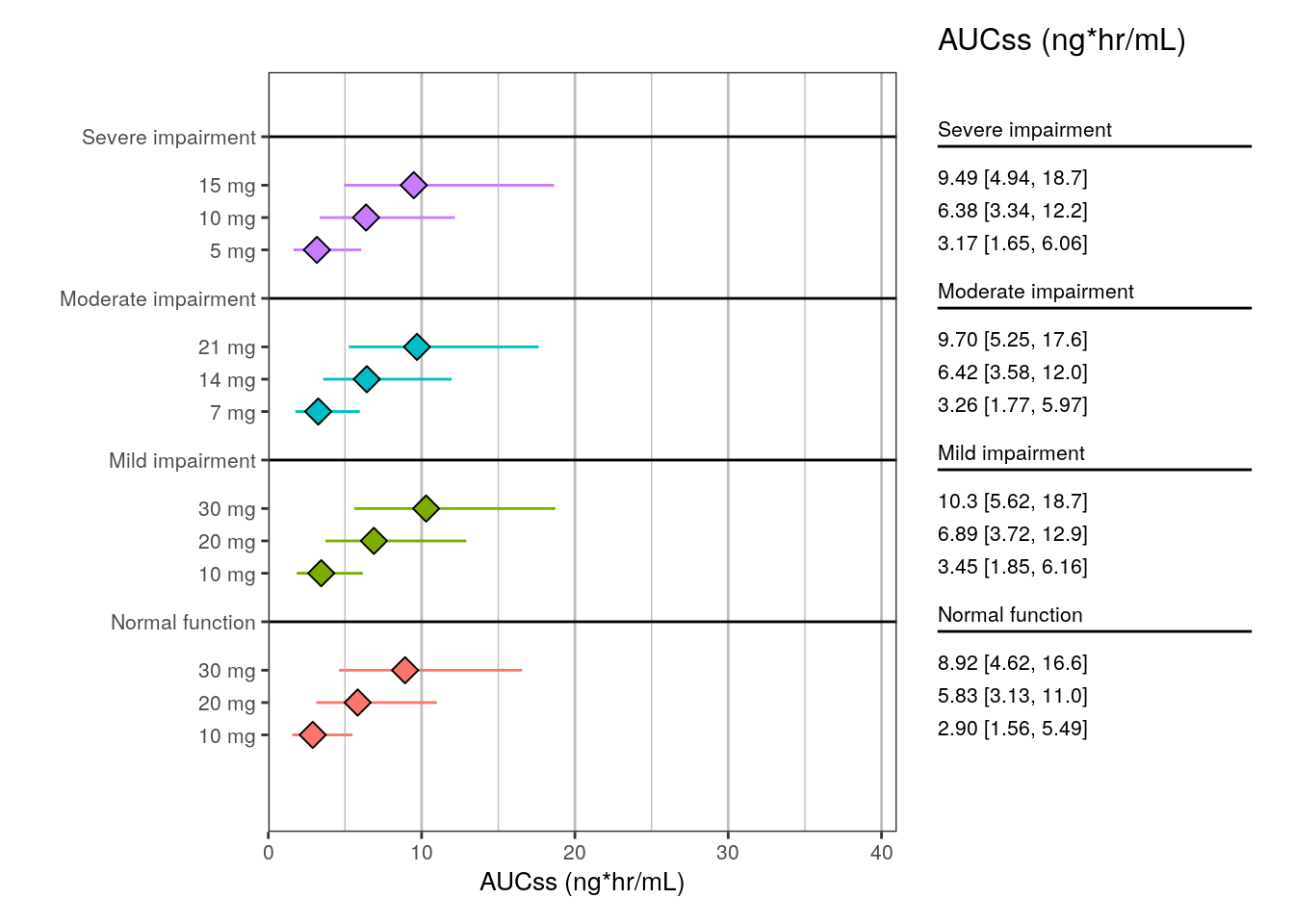
head(summ1)# A tibble: 6 × 5
group group_level mid lo hi
<fct> <fct> <dbl> <dbl> <dbl>
1 Normal function 10 mg 2.90 1.56 5.49
2 Normal function 20 mg 5.83 3.13 11.0
3 Normal function 30 mg 8.92 4.62 16.6
4 Mild impairment 10 mg 3.45 1.85 6.16
5 Mild impairment 20 mg 6.89 3.72 12.9
6 Mild impairment 30 mg 10.3 5.62 18.7 3.2 Step 2: create the forest plot
We create a plot of the summarized data using plot_forest(), passing in the summarized data and some additional settings.
plot_forest(
summ1,
vline_intercept = NULL,
x_lab = "AUCss (ng*hr/mL)",
x_limit = c(0, 41)
)
In the summarized data, the group column sets the outer grouping in the plot (renal function level) and the group_level column sets the levels within that grouping factor (in this case, the doses).
Also, note that the summary metrics are by default:
mid: the medianlo: the 5th percentilehi: the 95th percentile
All these summaries are customizable from the summarize_data() function.
We can make a similar plot for the original dosing scenario.
original <- filter(data, type=="original")
summ2 <- summarize_data(
data = original,
value = "AUC",
group = "RFf",
group_level = "dosef"
)
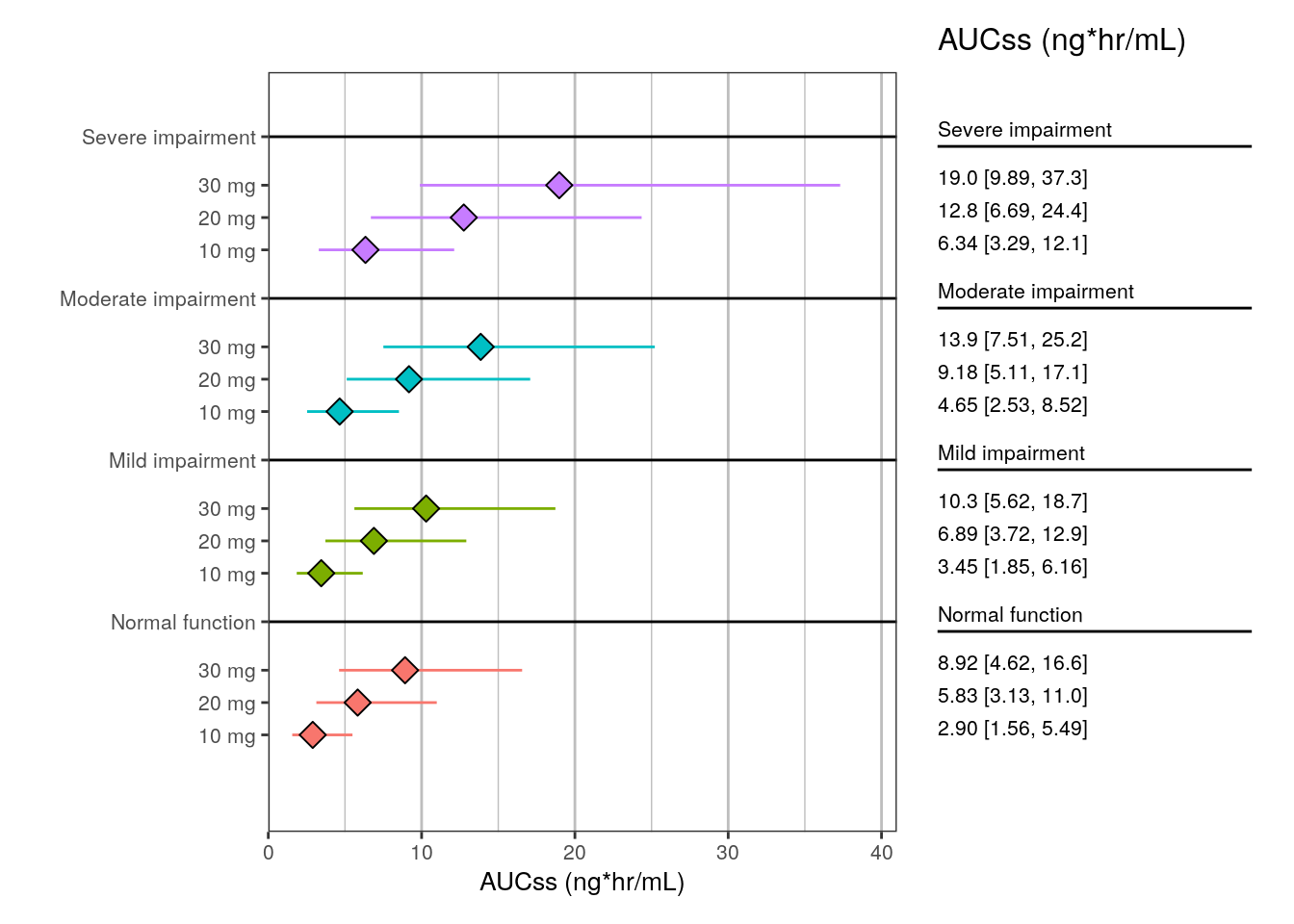
head(summ2)# A tibble: 6 × 5
group group_level mid lo hi
<fct> <fct> <dbl> <dbl> <dbl>
1 Normal function 10 mg 2.90 1.56 5.49
2 Normal function 20 mg 5.83 3.13 11.0
3 Normal function 30 mg 8.92 4.62 16.6
4 Mild impairment 10 mg 3.45 1.85 6.16
5 Mild impairment 20 mg 6.89 3.72 12.9
6 Mild impairment 30 mg 10.3 5.62 18.7 plot_forest(
summ2,
vline_intercept = NULL,
x_lab = "AUCss (ng*hr/mL)",
x_limit = c(0, 41)
)
3.3 Mark a region of interest
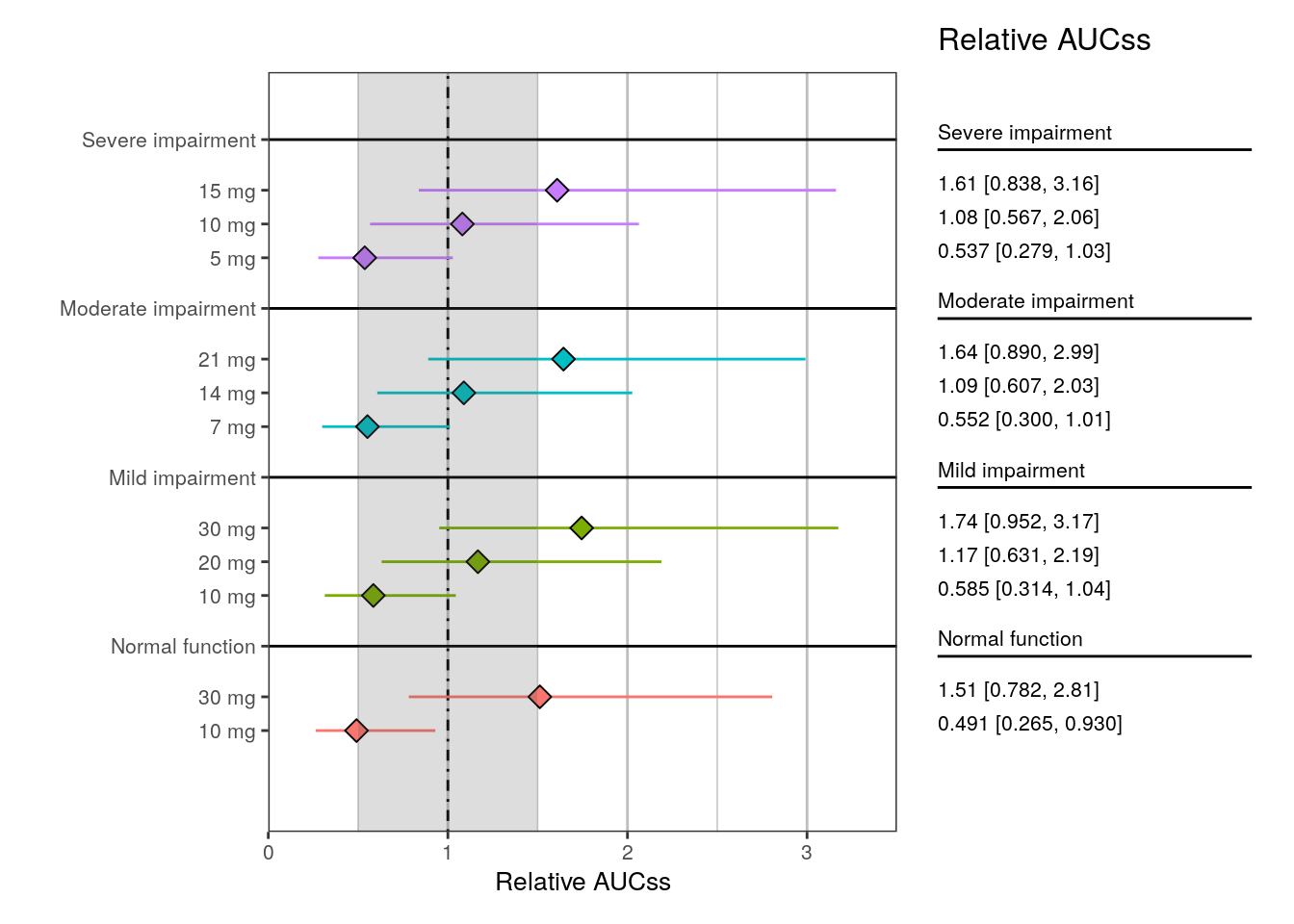
We can also annotate a region of interest or equivalence on the x-axis.
summ4 <- summarize_data(
data = adjusted,
value = "AUCrel",
group = "RFf",
group_level = "dosef"
)
plot_forest(
summ4,
vline_intercept = 1,
x_lab = "Relative AUCss",
x_limit = c(0, 3.5),
shaded_interval = c(0.5, 1.5),
shape_size = 3
)