library(here)
library(mrgsolve)
library(tidyverse)
library(rlang)
library(yspec)
library(pmforest)1 Introduction
This page demonstrates how to use the pmforest package along with mrgsolve to create covariate forest plots. These plots are used to provide context around estimated covariate effects by simulating model parameters (e.g., clearance) after varying the values of different covariates one at a time. The simulated parameters are normalized to parameters simulated at the reference covariate levels and all simulations are performed over the uncertainty distribution of the THETAs from the estimated model.
2 Tools used
3 Outline
This document is more involved than most of the other pages in this Expo.
First, we’ll load the
- data specification object
- relevant mrgsolve model
- data frame of bootstrap parameter estimates
Then we’ll use the mrgsolve model to simulate clearances across the bootstrap parameter estimate set for the reference covariate set.
Next, we’ll simulate clearances (again across the bootstrap parameter set) after varying the values of the different model covariates, one at a time. This illustrates how changes in the covariate translate into changes in clearance.
Finally, we’ll summarize the simulated clearance values at each covariate level and create a forest plot from these summarized simulations.
4 Required packages and options
5 Load the data specification object
We load the yspec object, which will provide information about the data set, including covariates in the model:
spec <- ys_load(here("data/derived/pk.yml"))We’ll modify the EGFR unit to be blank:
spec$EGFR$unit <- ""Then, extract a list of units to annotate the plot:
unit <-
spec %>%
ys_get_unit() %>%
as_tibble() %>%
pivot_longer(everything(), values_to = "unit")
head(unit)# A tibble: 6 × 2
name unit
<chr> <chr>
1 C ""
2 NUM ""
3 ID ""
4 TIME "hour"
5 SEQ ""
6 CMT "" 6 Read in bootstrap parameter estimates
We’ll retain only 200 rows for now, and we’ll number the rows.
post <- read_csv(here("data/boot/boot-106.csv"))
set.seed(10101)
post <- select(post, contains("THETA"))
post <- slice_sample(post, n = 200)
post <- mutate(post, iter = row_number())7 Read in the mrgsolve model
For the final model (run 106), we already have the simulation model coded up: 106.mod. This will enable us to simulate clearances from different covariate values.
mod <- mread(here("script/model/106.mod"), end = -1, outvars = "CL")
mod <- zero_re(mod)Note that we called zero_re() on the model object; this suppresses the simulation of random effects. In this task, we are only interested in simulated typical parameters.
8 Simulate
8.1 Simulate the reference
Simulate a dataframe of clearances at the reference covariate combination:
ref <- mrgsim(mod, idata = post) %>% select(iter = ID, ref = CL)
head(ref)# A tibble: 6 × 2
iter ref
<dbl> <dbl>
1 1 3.23
2 2 3.15
3 3 3.07
4 4 3.41
5 5 3.23
6 6 3.15Note that this simulation produces clearances at the reference covariate levels because we coded the default covariates to be the reference values in the mrgsolve model.
8.2 Simulate different covariate levels
Now, we have to simulate clearances after perturbing each covariate across levels of interest, one at a time.
To do this, we’ll create a named list of vectors, where the name reflects the covariate that is being perturbed, and the vector has all the values to examine.
x <- list(
WT = c(85, 55),
EGFR = c(Severe = 22.6, Moderate = 42.9, Mild = 74.9),
ALB = c(5, 3.25),
AGE = c(45, 20)
)For EGFR, we’ve included names for recoding the EGFR as a renal impairment stage (e.g., moderate renal impairment).
Now, we write a function that simulates each covariate at each of the prescribed levels:
ss <- function(values, col) {
COL <- sym(col)
idata <- tibble(!!COL := values, LVL = seq_along(values))
idata <- crossing(post, idata)
out <-
mod %>%
mrgsim_i(idata, carry_out = c(col, "LVL", "iter")) %>%
mutate(name = col, value = !!COL, !!COL := NULL) %>%
arrange(LVL)
if(is_named(values)) {
out <- mutate(
out,
value := factor(value, labels = names(values), levels=values)
)
} else {
out <- mutate(out, value = fct_inorder(as.character(value)))
}
out
}This function gets called usiung an imap variant to simulate clearances:
out <- imap_dfr(x, ss)
head(out)# A tibble: 6 × 7
ID time LVL iter CL name value
<dbl> <dbl> <dbl> <dbl> <dbl> <chr> <fct>
1 2 0 1 128 3.72 WT 85
2 4 0 1 104 3.49 WT 85
3 6 0 1 123 3.60 WT 85
4 8 0 1 18 3.64 WT 85
5 10 0 1 1 3.73 WT 85
6 12 0 1 59 3.70 WT 85 This function:
- Creates an
idata_setwith the current covariate name and values - Uses
tidyr::crossingto expandidatato include all the bootstrap samples we loaded frompost - Simulates the clearance
- After the simulation, we log the
nameof the covariate and thevalues which were simulated - Finally, we check to see if names were passed and use those to recode the covariate values
The result of the function is 200 clearances simulated at multiple levels of multiple covariates.
count(out, name, value)# A tibble: 9 × 3
name value n
<chr> <fct> <int>
1 AGE 45 200
2 AGE 20 200
3 ALB 5 200
4 ALB 3.25 200
5 EGFR Severe 200
6 EGFR Moderate 200
7 EGFR Mild 200
8 WT 85 200
9 WT 55 2009 Plot
9.1 Get ready to plot
Merge in the unit by name:
out <- left_join(out, unit, by = "name")
out <- mutate(out, cov_level = paste(value, unit))Merge in the reference clearances by iter and calculate the simulated clearance relative to the reference (relcl):
out <- left_join(out, ref, by = "iter")
out <- mutate(out, relcl = CL/ref)Set up labels and create a labeller object:
all_labels <- ys_get_short(spec, title_case = TRUE)
all_labels$EGFR <- 'Renal Function'
all_labels$CL <- 'CL (L/hr)'
plot_labels <- as_labeller(unlist(all_labels))9.2 Summarize the data
Call summarize_data from the pmforest package to generate summaries. We’re asking for the 2.5th and 97.5th percentiles of the clearance at each covariate level. summarize_data also gives us the median for plotting.
sum_data <- summarize_data(
data = out ,
value = "relcl",
group = "name",
group_level = "cov_level",
probs = c(0.025, 0.975),
statistic = "median"
)
head(sum_data)# A tibble: 6 × 5
group group_level mid lo hi
<chr> <chr> <dbl> <dbl> <dbl>
1 AGE "20 years" 1.03 0.957 1.10
2 AGE "45 years" 0.988 0.959 1.02
3 ALB "3.25 g/dL" 0.873 0.827 0.908
4 ALB "5 g/dL" 1.05 1.03 1.06
5 EGFR "Mild " 0.915 0.900 0.927
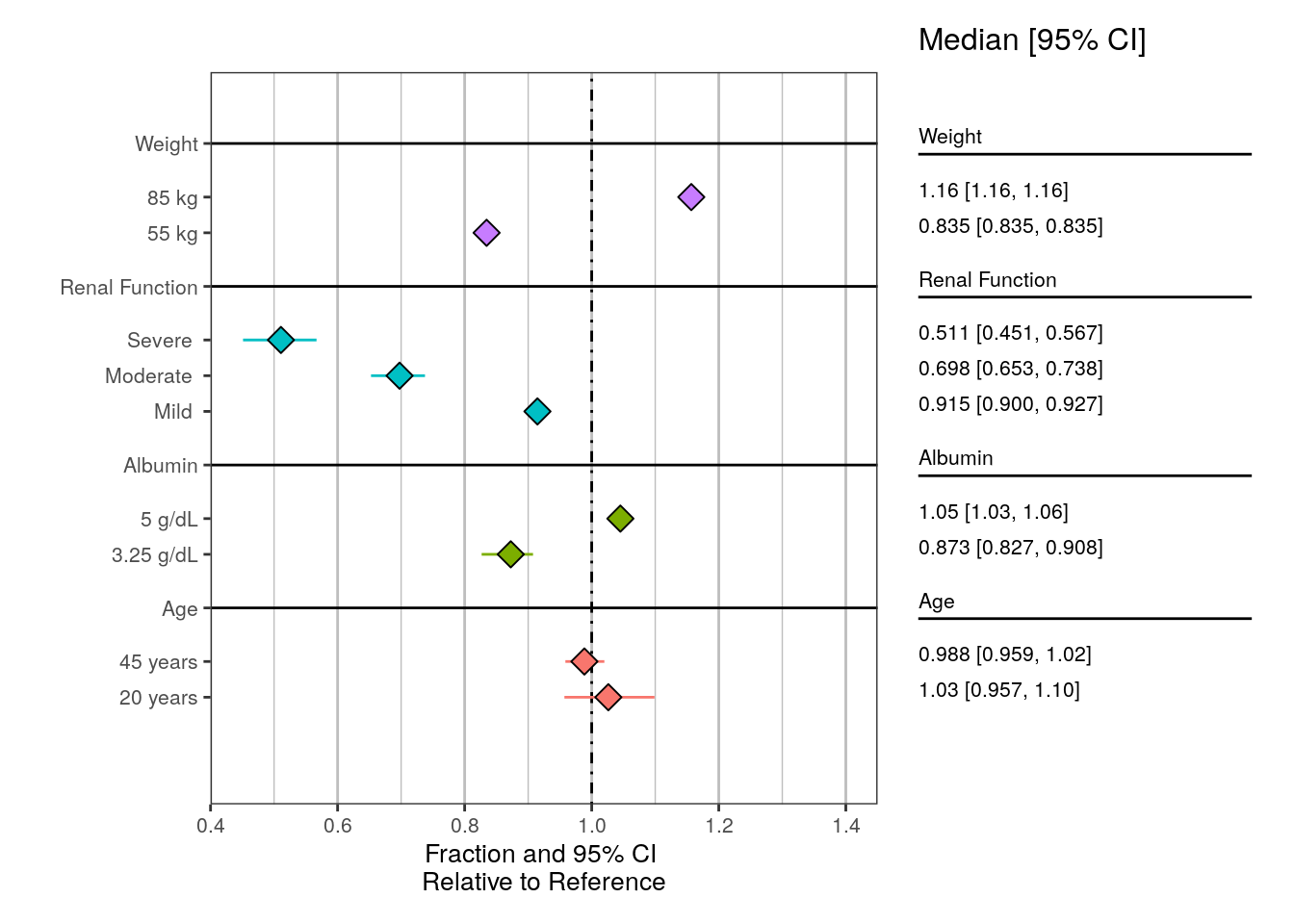
6 EGFR "Moderate " 0.698 0.653 0.7389.3 Create the plot
Please see the ?plot_forest documentation for details around each argument.
clp <- plot_forest(
data = sum_data,
summary_label = plot_labels,
text_size = 3.5,
vline_intercept = 1,
x_lab = "Fraction and 95% CI \nRelative to Reference",
CI_label = "Median [95% CI]",
plot_width = 8,
x_breaks = c(0.4,0.6, 0.8, 1, 1.2, 1.4,1.6),
x_limit = c(0.4,1.45),
annotate_CI = T,
nrow = 1
) clp