Introduction
One benefit of working with R Markdown is that you can reproduce an analysis at the click of a button. Parameterized reports extend this concept one step further and allow users to specify one or more parameters to customize their analysis. This is useful when creating diagnostic plots because you create your diagnostic plots once in an R Markdown template and then create those plots across multiple similar models.
Additional details on these kinds of reports are given in the Expo 1 page Parameterized Reports for Model Diagnostics. Here, we will simply point out the different structure of scripts and template that we use for NONMEM® Bayesian (Bayes) Model. (Figure 1).
This project includes three R Markdown templates that are rendered from two R scripts to create a summary html with floating table of contents and, in the case of the report-ready figures, annotate pdfs (Figure 1)
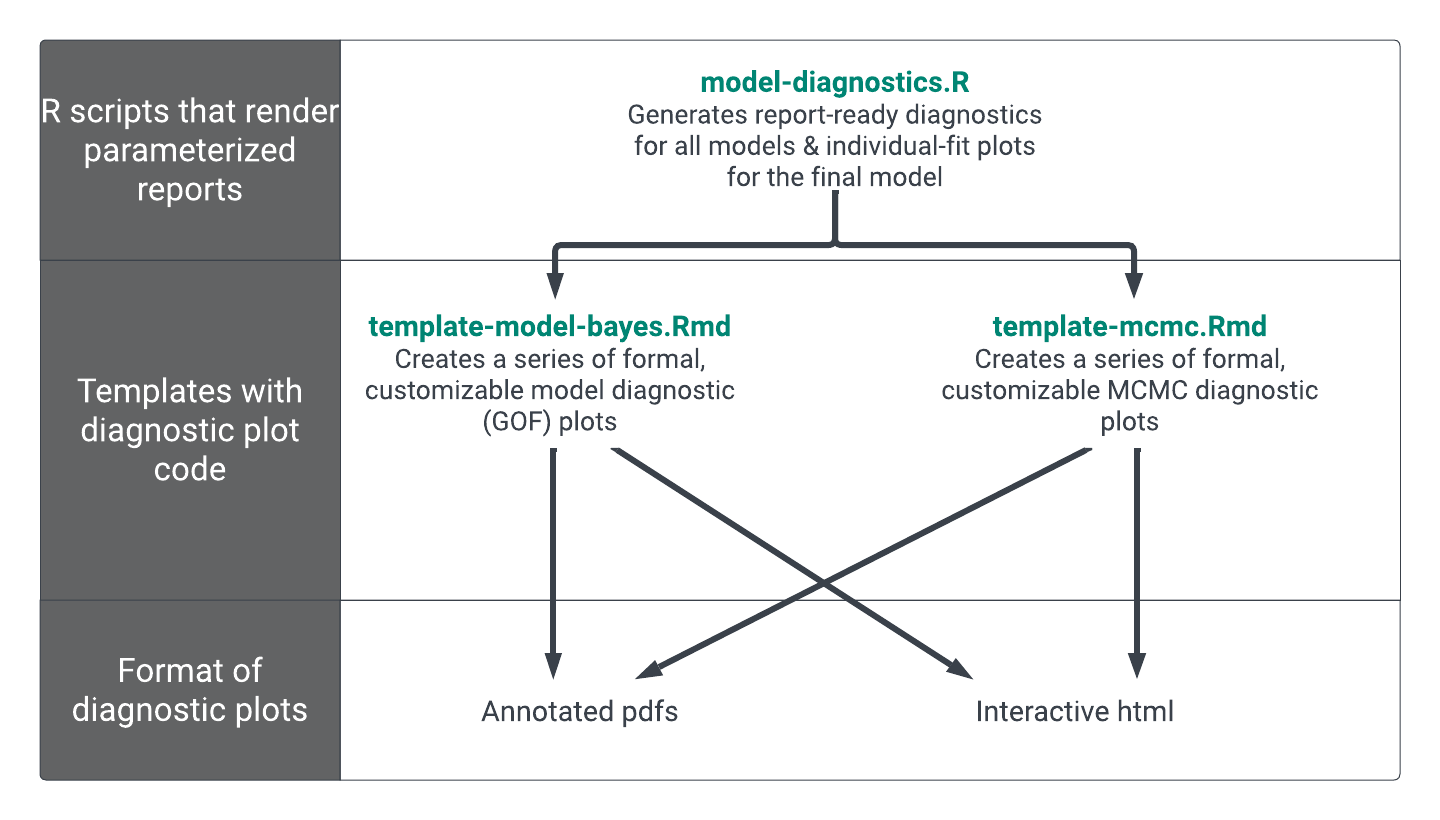
The output of these templates is flexible, here, we make individual .pdf and .png versions of the report-ready plots for use in our Latex reports. Each template also generates a single .html summary file too. We chose to render the summary file to an .html (rather than a .pdf) so that the rendered document includes a floating table of contents, allowing easy navigation through all the summaries and diagnostics generated in the template. We have included an example rendered model diagnostics report and an example rendered MCMC diagnostics report to show what this output might look like.
Additional information about parameterized reports can be found on the R Markdown: The Definitive Guide website.
Other resources
The following scripts from the GitHub repository involved in parameterized reports. If you’re interested running this code, visit the About the GitHub Repo page first.
- Script to render diagnostics templates: model-diagnostics.R
- Diagnostics Rmd templates: